Premium Only Content
Principle and Workflow of Whole Genome Bisulfite Sequencing - CD Genomics
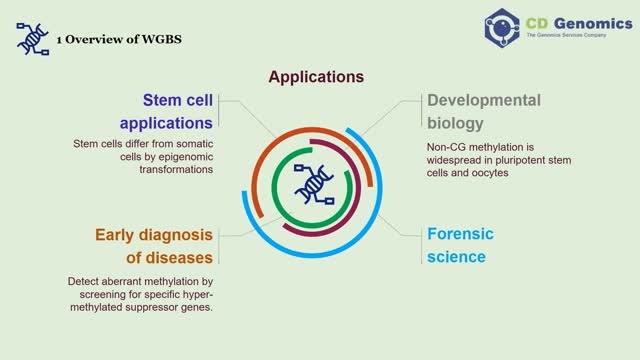
Epigenetic studies have confirmed that DNA-methylation modification of specific gene regions plays an important role in chromosome conformation and gene expression regulation. Methylation of DNA cytosine residues at the C5 (5meC) is a common epigenetic mark in many eukaryotes and is widely found in CpG or CpHpG (H=A, T, C). There are mainly three approaches, including endonuclease digestion, affinity enrichment, and bisulfite conversion (Table 1). Almost all sequence-specific DNA methylation analysis approaches require a methylation-dependent treatment before amplification or hybridization to maintain fidelity. Various molecular biology techniques, such as next-generation sequencing (NGS), are subsequently performed to detect 5meC residues. https://www.cd-genomics.com/principles-and-workflow-of-whole-genome-bisulfite-sequencing.html
-
 12:49
12:49
kikoooo
4 years agoMicrobial Genomics
22 -
 15:48
15:48
dorawest111
4 years agoProtein Sequencing
25 -
 4:55
4:55
KTNV
4 years agoWhole House Clean
2653 -
 13:56
13:56
What Is Truth
4 years ago $0.01 earnedPS16 Heisenberg Uncertainty Principle
41 -
 15:55
15:55
ferrel
4 years agoWriting, The Werewolf Principle
20 -
 0:21
0:21
WFTX
4 years agoWhole Foods turkey warning
101 -
 0:30
0:30
KJRH
4 years agoWho's Hiring: Whole Foods
591 -
 2:13
2:13
WKBW
4 years agoSchool House 7 - Sequencing
18 -
 9:51
9:51
AFV
4 years agoWhole Lotta Fails
4.13K1 -
 48:36
48:36
PMG
1 day ago $0.06 earned"Parkland Parent Speaks Out On Kamala Harris Using Victims"
4172